MRSA: scientists on trail of antibiotic resistance
Whole genome sequencing allows researchers to monitor how resistant bacteria travel from humans to humans, animals to humans and from humans to animals.
Methicillin-resistant Staphylococcus aureus (MRSA) is one of many antibiotic-resistant bacteria that pose a threat to public health.
It is for instance involved in a number of inflammatory diseases that are difficult to cure.
MRSA has the ability to almost indiscriminately jump from humans to animals and back again, which makes it hard to monitor its infection activities.
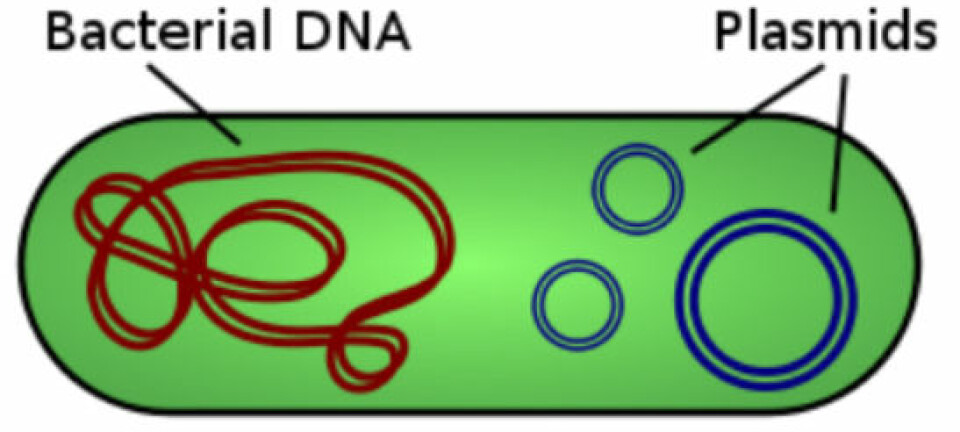
Now researchers, including a team from the Danish State Serum Institute (SSI), have used whole genome sequencing (see Factbox) to more accurately map the spread of MRSA infection from animals to humans.
This may in the long run make it easier to prevent future dissemination of resistant bacteria.
”With whole genome sequencing, we will get a clearer understanding of bacterial infection routes. It will provide us with a more intelligent way of fighting and preventing the spread of resistance bacteria,” says one of the researchers behind the new study, Robert Leo Skov, MD, of the SSI.
The study is published in The EMBO Journal.
Resistance in Danish livestock
In the study, the researchers used whole genome sequencing of MRSA found in two Danish livestock populations that had infected wounds.
The aim was to use whole genome sequencing to determine:
- Whether it was the same strain of bacteria that had infected both of the livestock populations.
- The infection conditions between animals and humans.
The sequencing revealed that the two populations had been infected by different strains of bacteria, and also that the humans on each of the two farms had inflammations caused by the same MRSA that was found in the animals.
Based on the number of mutations in the genome, the researchers could also conclude that the route of infection was from animals to humans and not the other way round.
”Here, whole genome sequencing provides us with further evidence that interaction between animals and humans can cause a spread of resistant bacteria,” says Skov.
A special MRSA
The MRSA the researchers looked at in this study is rather interesting in itself. These bacteria contain a gene for antibiotic resistance, known as ’mecC’.
This gene, which was only recently described, gives the researchers a great opportunity to learn more about the spread of MRSA.
”MRSA with mecC is not very common, neither in Denmark nor in the rest of the world. It therefore presents us with the opportunity to monitor the spread of this type of MRSA and take notes whenever we find it,” says the researcher.
“It gives us a unique view of the development of the spread of a disease. We can learn a great deal from that.”
The researchers can also go back in time and see how MRSA with mecC has existed in Denmark at least since 1975, when it was found in a Danish patient with bacteremia.
-------------------------
Read the Danish version of this article at videnskab.dk
Translated by: Dann Vinther