Researchers' Zone:

These magic bullet-proteins are revolutionizing modern medicine
A treatment for snake venom, COVID-19 tests and cure for autoimmune disease and cancer. The possibilities of molecular binders are manifold.
Did you know that a certain type of molecules is not only vitally important for your immune system to fight infections, they are at the same time the basis of the diagnostic antigen tests used during COVID-19?
And not only that: They are also the active ingredient in snake antivenom, and they are currently part of a revolution in modern medicine.
These molecules are called ‘molecular binders’. You have probably already heard of one kind of molecular binder, namely antibodies.
In this first part of a two-part article series, we will explain what they are, what they are used for – and why they are called ‘magic bullets’.
Molecular binders – the magic bullets of the microscopic world
A molecular binder is a protein (or another type of molecule like small molecule) that binds to another protein like two pieces of a jigsaw puzzle.
One of the best-known molecular binders is antibodies, which are proteins produced by the immune system that bind, for example, intruding bacteria or viruses and help fight infections.
Antibodies have often been referred to as ‘magic bullets’ as they target and kill intruding organisms while not harming the body.
The magic of antibodies arises from their ability to bind very specifically to a target while not binding to anything else – a concept known as specific binding.
This ‘magic’ ability, although it is invented by nature, has been exploited by human ingenuity for the design and engineering of antibodies with tailored properties.
However, to exploit such a valuable trait, one needs to understand it first. Thus, the origin and mechanisms of antibody specificity have been heavily explored.
It was found that the specific ‘binding’ of a binder to another protein is primarily due to shape complementarity. That basically means that the protein fits together with its target nicely.
Indeed, proteins are large (in a microscopic sense) amino acid chains that have a particular three-dimensional shape.
This shape can be sphere-like, elongated, contain grooves, or extruding ‘loops’. By virtue of this shape, a molecular binder might be complementary to its target, just like the groove and cleft of the jigsaw puzzle pieces.
But why are these binders so useful?
Molecular binders are commonly used for three different purposes. These uses are based on their ability to bind to predefined targets and not to other proteins.
1. Can identify proteins
One of the main uses is as laboratory reagents used for the identification and quantification of proteins.
One example of how binders can be used as laboratory reagents is for identifying if a specific protein of interest is present.
Finding a specific protein can be done by using a colour-labelled binder towards that protein. The colour-labelled binder that has attached itself to the protein in question can then be spotted on microscopy images. In this way scientists can classify different cell types in microscopy images.
This is a powerful technique used in many branches of science from cancer research to plant science.
2. Can detect corona virus and other diseases
In fact, the COVID-19 ‘lateral flow test’ (a.k.a. quick test or antigen test) uses antibodies as binders specific to the coronavirus.
The quick test has two lines on it; the ‘test line’ and the ‘control line’. The antibodies specific to the coronavirus are located on the ‘test line’.
The sample is first exposed to antibodies carrying a colour label such that any coronavirus proteins in the sample get coloured. The coloured corona proteins then flow though the strip and form a coloured line as they bind to the antibodies on the test line.
3. Can push the right buttons
Molecular binders’ arguably most important function, however, falls within modern medicine as therapeutics.
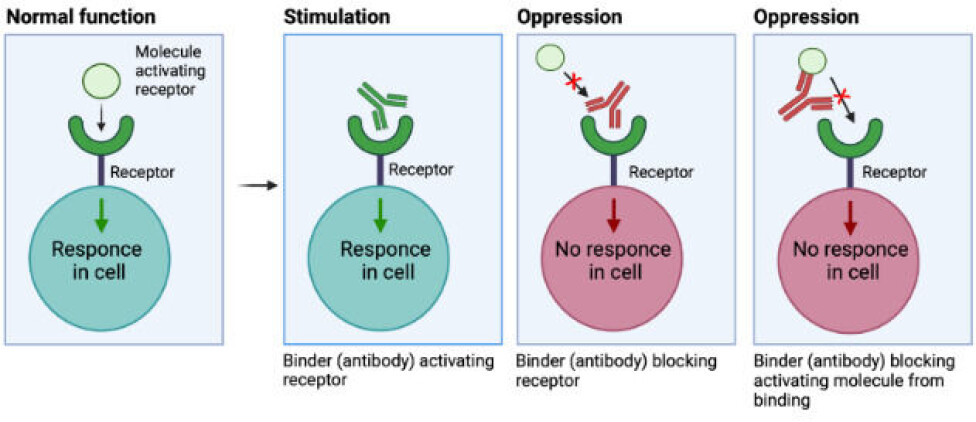
We can, for instance, exploit their ability to bind to receptors on cells and activate or deactivate processes, which are key in certain diseases.
There are different principles governing the interaction between the binder and the receptor.
The binder can be seen as ‘pushing the right button’, similar to the natively binding molecule, mimicking its function.
World famous molecular binder: insulin
As an example of this, we can take one of the most well-known hormones, insulin.
Among many other functions, insulin binds to receptors on cells and mediates the uptake of glucose into the cells. We can imagine how a binder might bind to the same receptor and mimic insulin.
The result will be glucose uptake but without the addition of insulin. Although binders will probably not outcompete modern diabetes drugs, this example shows you the concept of how binders can be used in treating disease.
The binder may also oppress the signalling of a receptor by blocking access of the activating molecule (like hormones) to the receptor by binding to either the receptor or the activating molecule, as seen in the figure above.
One example of is the drug Omalizumab used against Asthma because it binds to a type of signalling molecules in the immune system, which is excessively present in this disease.
A revolution
This ability to manipulate cellular responses makes binders extremely powerful.
It is no less than revolutionizing, because the methods for developing binders have advanced significantly in the last decades.
This means that once you understand what the cause of a disease is at the receptor level, it is relatively straight forward to develop potential drugs right away.
In addition, proteins are native to the body and its metabolic pathways, meaning that protein-based drugs are appealing compared to small molecule drugs (like paracetamol or acetylsalicylic acid). Small molecule drugs can accumulate or place strain on the body in the breakdown process. This can be avoided by replacing them with molecular binders.
Difficult snake bite venoms can be treated
However, the potential of these magic bullets does not stop there.
More recently, specific binders (antibodies) have also been explored in treating snakebites. Snake venoms consist of cocktails of toxic proteins, which employ different mechanisms for manipulating important receptors or destroying the cells and organs in your body.
If a molecular binder binds to one of these toxic proteins, it can potentially prevent the toxin from exerting its toxic effects.
For example, some snake venom toxins bind receptors in the nervous system, causing muscle cramping. If a binder is attached to this type of toxin before it reaches the receptor, this will prevent the toxin from binding to the receptor and therefore neutralise it (analogous to what is seen to the right in figure 1).
In this way, binders which decrease the lethality of snake venom in mice have been successfully discovered. Clearly, these magic bullets have many interesting uses, but how does one go about discovering binders?
The key here is ‘directed evolution’ in combination with ‘protein libraries’.
For now, we will look at protein libraries, and reserve the Nobel prize-winning technology of ‘directed evolution’ for the upcoming second article.
Protein libraries, the treasure troves of protein binders
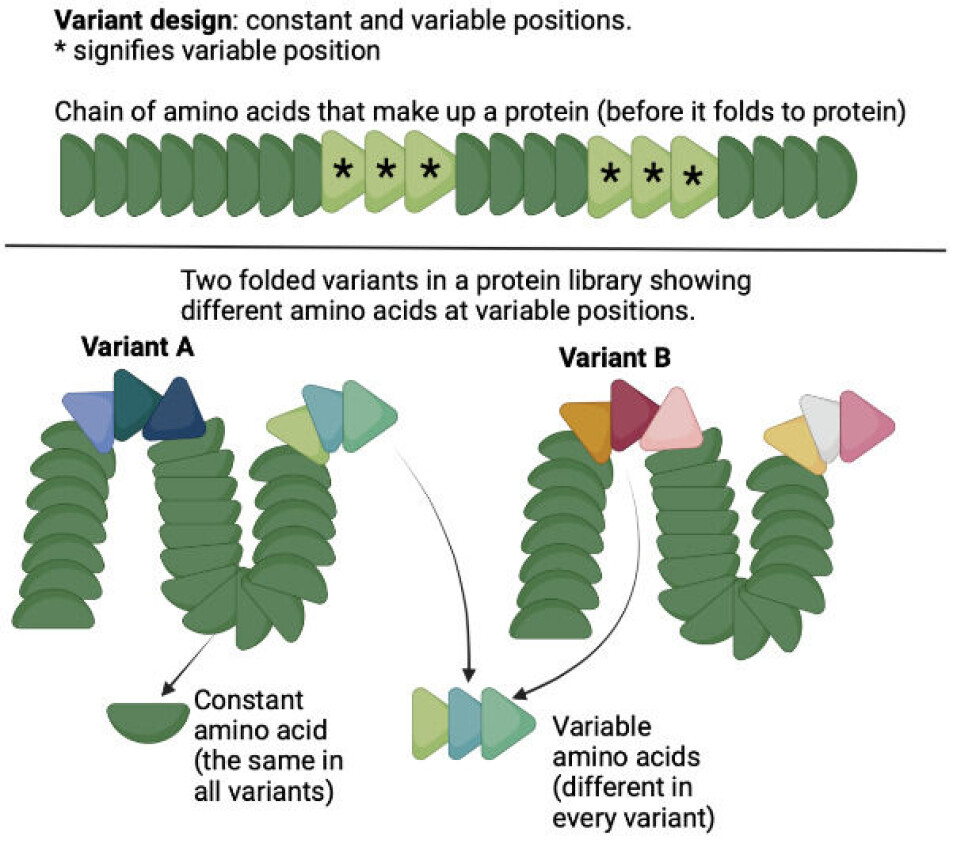
Simply put, a protein library is a large collection of slightly varying proteins.
Such a library typically consists of a huge population of up to a billion slightly different proteins (called variants), all having the same overall structure but each varying slightly from one to another.
A bit like how all books in a library are similar (in size, shape, materials etc.) but still different.
However, the origin of proteins produced in the library can either be (1) taken from nature or (2) be made by human engineering.
Animal origin or engineered?
In the first case the proteins are antibodies produced by animals. The animals have been given the target (snake venom or a disease for example) and have thus developed antibodies.
All of these antibodies share their overall structure, but differ slightly in some areas, that gives the binding abilities.
Alternatively, the protein in the library can be engineered, by taking a protein structure and introducing random variations creating different kinds of the original proteins (this is called naïve library).
A very important point here is that the variants are not entirely random!
All the variants in a library contain the same backbone structure, and only a few carefully chosen ‘slots’ in the sequence are allowed to vary and contain any of the jigsaw pieces available in the pool of 20 different kinds of pieces.
This can be seen in figure 2 below. Given that the variable amino acids can be combined in a vast number of ways, most variants are slightly different from the rest of the ‘library members’.
One protein for every human on earth
If the protein library is of high quality and diversity, which means that it includes a substantial number (For example a library can include >1010 variants which is more than there are humans on earth) of different and functional binders, there will be some binders against any possible target of interest.
This is the case, even if the target has never existed in nature.
This means that if a new disease comes to earth on a comet, binders could also be found in the library against targets from the comet (although the target must not be too small because proteins are rather large)! These binders are present by mere chance owing to the large number of combinations present.
Nevertheless, the majority of variants will not be binders towards a given target.
So once a good quality library is in place, it is now possible to search these libraries and discover the few binders amongst a pool of non-binders.
You have now learned about the wonders of molecular binders, but an unanswered question remains: From the protein library containing billions of proteins that are too small to be seen, how do you select the few proteins that are binders?
Indeed, this requires Nobel prize-winning technology, directed evolution and molecular display – a concept that we explore in the next article.
This article was originally published on our Danish sister site, Forskerzonen.
References
- Andreas Sixsten Hallstein Rygaard's profile (Linkedin)
- Anne Ljungars' profile (DTU)
- Thomas Fryer's profile (ResearchGate)
- Timothy Patrick Jenkins' profile (DTU)
- 'Designing Repeat Proteins: Well-expressed, Soluble and Stable Proteins from Combinatorial Libraries of Consensus Ankyrin Repeat Proteins', JMB (2003), DOI: 10.1016/S0022-2836(03)00896-9
- 'Antivenom for Neuromuscular Paralysis Resulting From Snake Envenoming', Toxins (2017), DOI: 10.3390/toxins904014
- 'In vitro discovery of a human monoclonal antibody that neutralizes lethality of cobra snake venom', mAbs (2022), DOI: 10.1080/19420862.2022.2085536





