Are all cancer cells the same?
Not necessarily. And being able to identify the differences at a genetic level could lead to more effective, personalised treatments.
Take two cancer cells and compare their genomes. Surprisingly, they can be quite different.
This genetic variation is one of the hallmarks of cancer, and one reason why treating cancer is so hard.
If a tumour is made up of cells with many different genomes, a single drug might not kill them all. But, knowing the genetic variation can help us develop targeted treatments.
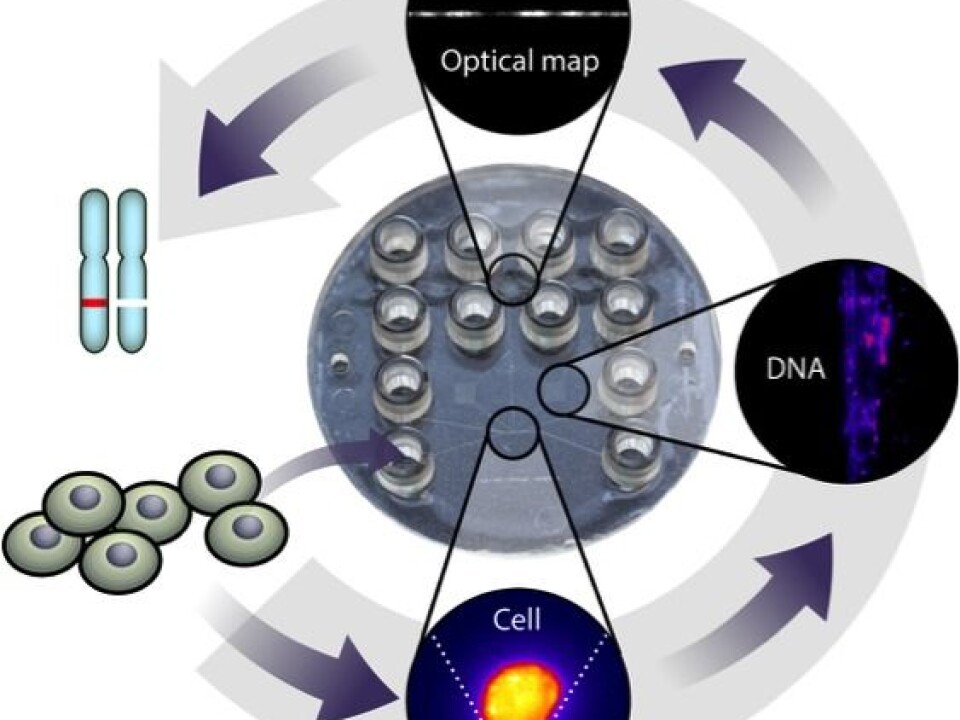
As part of our research, we have trapped a single cancer cell on a plastic device (see figure), extracted its DNA, and produced a coarse grained picture of this DNA’s sequence.
Our results are published in the journal ‘Proceeding of the National Academy of Sciences of the United States of America’ (PNAS).
Optical mapping of DNA from a single cell
We used a technique called optical mapping which provides large-scale information about the genome.
It works like a map of the world with forests, lakes, and mountains, but without the finer details like roads, houses, and small cities.
By comparing optical maps from a single cell with a reference map for the average human cell, we can pinpoint differences between them. This information can help to uncover genomic heterogeneity within a tumour and may even indicate how cells have developed into tumours.
Optical mapping of DNA from a single cell consists of four steps:
• First we capture a cell and extract long fragments of its DNA.
• Then we dye the DNA fragments with a fluorescent dye.
• Afterwards we heat the DNA molecules. The dye sticks better in some places than others, depending on the DNA sequence. This leaves a barcode-like pattern on the molecules.
• Finally we stretch and image the molecules under a microscope to read the ‘barcodes’.
We developed a low-cost plastic device that integrates all four steps. The steps are shown in the figure below.
The ‘barcode’ works like a fingerprint: It identifies which part of the reference genome a DNA molecule is closely related to. And it can even reveal differences between the imaged DNA molecule and the reference genome.
Sequencing versus optical mapping
So why use optical mapping and not just the usual DNA sequencing that you may already be familiar with?
The usual DNA sequencing methods have the advantage that they have single-base-pair resolution, meaning that every base pair in a DNA molecule can be identified. Given enough material, one can sequence the entire genome from a human cell – approximately six billion bases – in a few days.
But it is challenging to sequence a single copy of a human genome. And that is all we have when we start with a single cell.
One of the challenges is that the usual DNA sequencing methods require multiple copies of the genome. Since there is only one copy of the genome in each cell, the first step is to copy the genome multiple times. This is called DNA amplification and is routine chemistry. But copying errors occur occasionally, which obscure the results.
Another challenge is that each copy of the DNA molecule is randomly cut into smaller pieces, just a few hundred base pairs long. These pieces are then sequenced. The results, the so-called ‘reads’, are then assembled into a full genome by matching partially overlapping reads.
It is also very difficult to detect structural variations, such as repeating patterns or inserted/missing genomic elements that are longer than the few hundred-base-pair read lengths. But it is just this type of structural information that may prove useful for the selection of cancer therapies.
There is, at least in theory, a more obvious and efficient way to read the DNA sequence. The genome is coded on 48 DNA molecules, in two nanometres thick fibres, up to eight centimetres long. So, why not just read the DNA sequence from one end to the other?
Optical mapping almost does this: It provides a rough ‘fingerprint’ of the underlying DNA sequence of DNA molecules up to one million base pairs long. That is, much longer than the short read lengths of DNA sequencing. And optical mapping also avoids the amplification steps needed for DNA sequencing.
The long fragments make it possible to detect structural variations that are from a few kilo-base pairs up to several 100 kilo-base pairs. Smaller variations can be detected with DNA sequencing.
So, the two methods (sequencing and mapping) are complementary. And the same DNA molecule can, in principle, be both optically mapped and then sequenced.
Towards a more efficient and personalised cancer treatment
The ability to sequence the genome at the single-cell level could lead to more efficient and individualised cancer treatments. But as we explained above sequencing the DNA from a single-cell is still demanding using current sequencing techniques.
We have demonstrated for the first time that optical mapping can detect large-scale genetic variations in a DNA molecule taken from a single cell. And we do it without the amplification steps required by DNA sequencing methods.
The sample preparation was done entirely on a single-use lab-on-a-chip device, starting from a single cell and finishing with useful genomics data.
This is an important technological aspect of our work, as the device reduces the use of expensive chemicals and minimises the risk of contamination.
To be clear, our work is still in the research phase. Our device is not yet ready for use in hospitals, and the challenge now is to increase the throughput, so we can analyse more DNA molecules at a time. The goal is to map all the DNA from a single cell.





