Researchers' Zone:

How a Nobel prize-winning technology is speeding up evolution
What if we could test billions of possible cures for a disease almost in an instant? That is what directed evolution does – and it is revolutionizing modern medicine.
In 2018 the Nobel prize in chemistry was awarded to three researchers: George P. Smith, Gregory P. Winter, and Frances H. Arnold for their invention of ‘directed evolution’.
Since its invention, it has revolutionised drug discovery, has been used to discover the world’s best-selling drug, and, in addition, has been used for the discovery of molecules crucial for diagnostic tests for many diseases.
In this second article of a two-part series, we will explain a protein engineering technique known as directed evolution, what it is, and how it is used to discover molecular binders. In the first article we explained what molecular binders are and what they can be used for.
The origin of directed evolution
Molecular binders are proteins with a shape that allows them to bind specifically to other proteins, which is immensely useful in science.
But how can the discovery of a molecular binder with a complementary shape towards a target be conducted?
This is where directed evolution presented a disruptive solution: It is a sped-up version of many generations of evolution.
In directed evolution, a certain ‘goal’ is enforced by the scientist by applying a so called ‘selection pressure’. It is a way of selecting for a certain trait.
As an example, arguably the first instance of directed evolution is crop-breeding. It is the process, by which humans select plant individuals for breeding based on certain favourable traits and, because of this, enhance these characteristics in a future population.
Another use of directed evolution is for the discovery of molecular binders, which is the focus of this article.
In directed evolution for binder discovery, the ‘goal’ of selection is not favourable plant traits, but strong binding to a specific target.
This scientific field has its roots in experiments performed in the late 1960s, but gained momentum in the 80s and 90s with the advent of a technique called ‘phage display’.
Directed evolution for binder discovery can be done in a small plastic tube using a library of a billion different protein variants – what we called a ‘protein library’ in our previous article.
First find, then amplify
You can use directed evolution to select the protein-variants in the ‘protein library’ that bind (binders) from those that do not bind (non-binders).
Conceptually, this can be thought of as testing (displaying) every single variant in the library against the target. Testing if the protein-variant binds and flushing away the ones that do not bind as it is seen in figure 1 (for further explanation, see below).
Once you have washed away the non-binders and are left with the binders, you need to identify and amplify them.
Two problems need to be solved, before this can be done:
Firstly, the binders are proteins and for that reason, they are hard to sequence (proteins are made up of chains of amino acids and when you sequence them, you determine the order of the amino acids in the chain. See fact box for more on sequencing).
Secondly, the binders cannot be amplified directly.
The genotype phenotype link
These two issues are elegantly solved by linking genotype and phenotype. Genotype and phenotype are biology terms that can be understood in terms of Lego. The genotype is the instruction, and the phenotype is the built Lego structure.
In biology, the genotype is the instructions in the DNA code and the phenotype is the proteins which are made from these instructions. Unlike protein, DNA is easy to sequence and amplify. Thus, the two issues described above are solved by linking the proteins to their DNA blueprint.
The DNA can be easily sequenced (that is, finding the order of genetic letters).
This can be thought of as putting the building instructions each variant directly on all the different variants so that once the binding variants are isolated and identified, they can easily be built again (instead of having to re-build it from looking at it).
To achieve the link, several technologies have been developed, such as phage or yeast display (more below).
To sum up: We now have a library of variants, where all are slightly different versions of the same overall structure. These variants are each connected to the exact DNA that carries the building instructions for that variant.
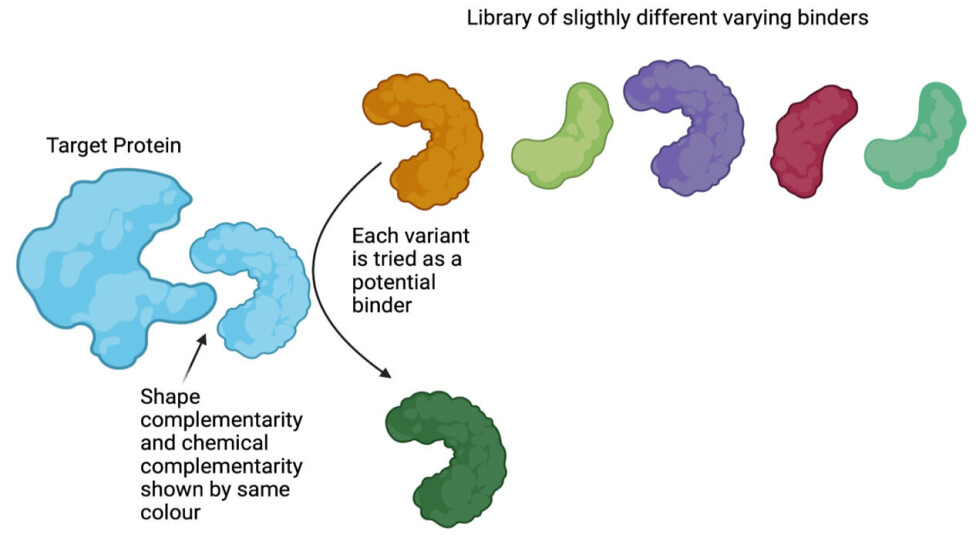
Discovering binders against targets of interest by molecular display
So how can we separate the billions of variants linked to DNA based on whether the variant binds to the target in a small plastic tube?
The key here is ‘fixing’ the target with the binders in a way that allows them to be separated from the non-binding proteins and everything else.
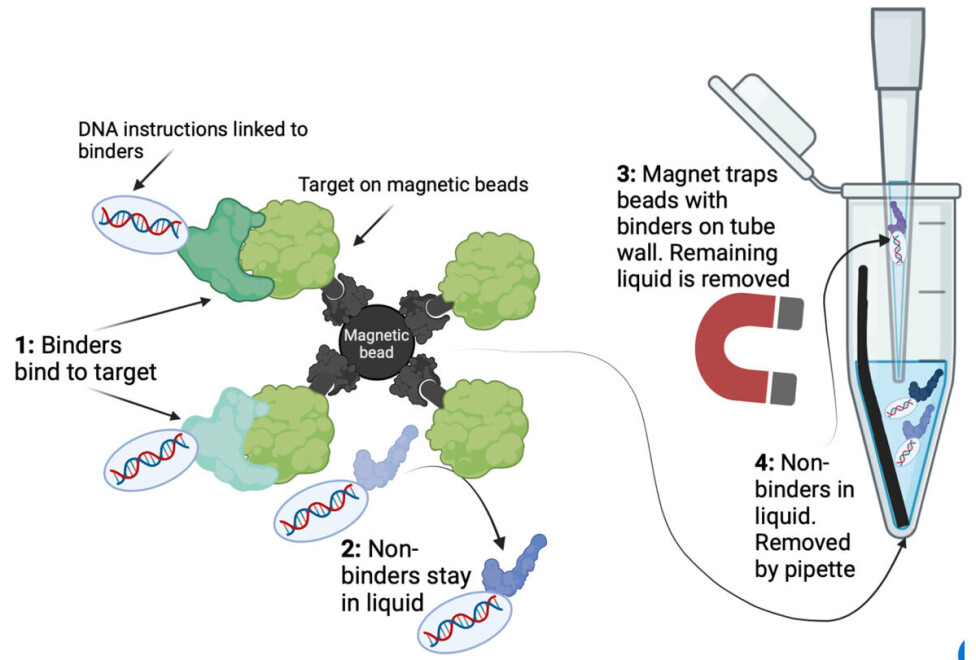
One way to achieve this is to attach your target to microscopic magnetic beads.
The smart thing about magnetic beads is that if you put a magnet next to the tube, all the beads with your binders will gather on the side of the tube. The remaining liquid with all non-binders can be discarded using a pipette.
As seen on figure 2, the binders indirectly stick to the beads because the beads are linked to the target protein.
In this way, binders and non-binders are separated, as the liquid mainly contains non-binders, and the binders are stuck on the target on the magnetic beads.
This can be seen in figure 2. Repeating this selection yields increasingly strong binders and is called ‘iterative bio panning’.
Yeast surface display
The linking of the DNA instructions to the protein variant (phenotype-genotype link) is a technical challenge which has been solved in different elegant ways.
The first discovered and most well-studied method is called ‘phage-display’ (read more about the method here); however the focus of this article falls on ‘yeast surface display’.
In this method, each one of the protein variants of the library is put on the surface of a yeast cell and inside the cell is the DNA instruction for the specific variant (a bit like the spike proteins on the corona virus that we have all seen in drawings).
By doing this the isolation of binders from non-binders is done using magnetic beads as described above. But as it turns out, another very cool way of isolating binders is possible with yeast surface display.
The most significant advantage of yeast surface display is that it allows for single-cell sorting based on the binding strength of each variant. Now what does that mean?
»Next please« – Sorting one billion cells one by one
The single-cell sorting is accomplished by a process called Fluorescence-Activated Cell Sorting (FACS).
The central thing in a FACS experiment, is a large collection of yeast cells, each expressing one single library variant. That is one of the many proteins in the library – like the ones you see in the top of figure 1.
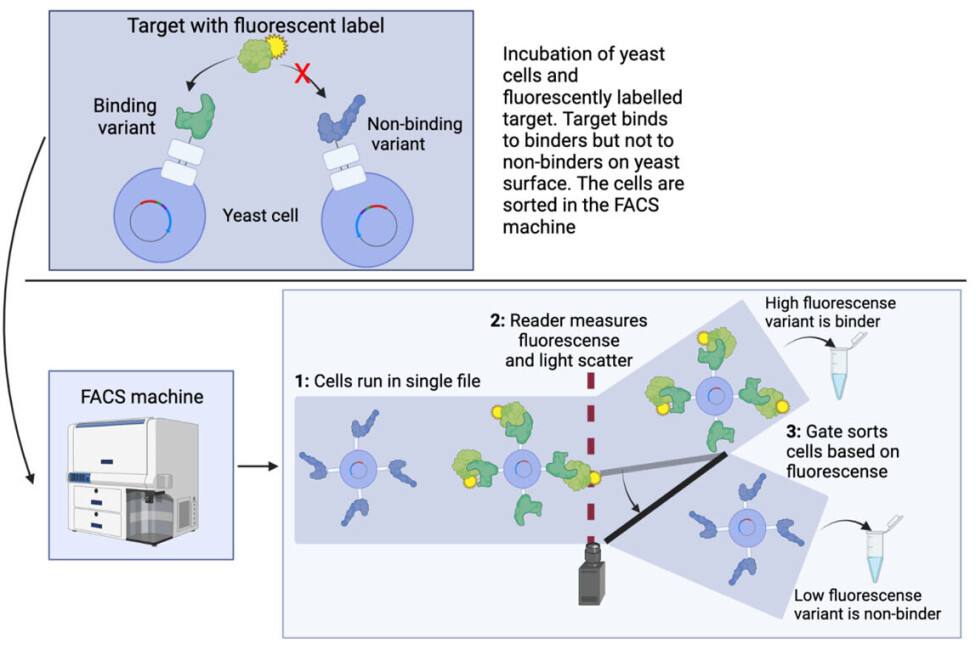
This collection of cells is mixed with the target protein that is coupled to a fluorescent dye (meaning that it glows when a light is shined on it) as seen in the top of figure 3.
When mixing fluorescent targets with yeast cells, the fluorescent target proteins will only bind on the surface of the yeast cells carrying a variant which is a binder, making these yeast cells fluorescent.
Now comes the technological wonder of the FACS machine: All yeast cells are run in a small tube, such that they come in a file with only one cell at a time.
The fluorescence of each cell is measured and corresponds to the amount of labelled target. The stronger the binding is, the more target is bound and the brighter the fluorescent colour.
The cells are then sorted one by one based on fluorescence intensity (remember, there are millions of cells). This can be seen in the bottom of figure 3. In this way, not only can the binders be isolated from the non-binders, but an indication of the strength of the binder is also directly obtained based on the strength of the fluorescent signal.
This allows for rapid and precise identification of molecular binders in a very short time (within weeks) and can rapidly accelerate drug discovery.
This is especially beneficial when speed is key, such as during a pandemic.
The world’s best-selling drug ever
Being able to discover binders is extremely powerful, as it allows for interaction with and manipulation of almost any biological system and, therefore, can be used in almost any pharmaceutical context for treating diseases.
Indeed, since its inception, directed evolution has revolutionised drug discovery.
We are now able to screen billions of antibodies against any disease of interest, including, cancer, tuberculosis, HIV, COVID-19, and autoimmune diseases.
Breakthroughs in developing better and cheaper snake antivenoms have also recently been made using directed evolution and further endeavours in this field are being pursued.
In fact, directed evolution has already proved its potential as it led to the discovery of the best-selling drug in the pharmaceutical history, Humira (more than 200 billion dollars), which is used to treat a plethora of different autoimmune conditions.
Overall, directed evolution is an extremely powerful technique, and we have certainly not seen the last of this cutting-edge technology.
This article was originally published in Danish on our sister site, Forskerzonen.
References
- Andreas Sixsten Hallstein Rygaard's profile (Linkedin)
- Anne Ljungars' profile (DTU)
- Thomas Fryer's profile (ResearchGate)
- Timothy Patrick Jenkins' profile (DTU)
- 'In vitro discovery of a human monoclonal antibody that neutralizes lethality of cobra snake venom', mAbs (2022), DOI: 10.1080/19420862.2022.2085536
- 'Basics of Antibody Phage Display Technology', toxins (2018), DOI: 10.3390/toxins10060236
- 'Recent Advances in Novel Lateral Flow Technologies for Detection of COVID-19', Biosensors (2021), DOI: 10.3390/bios11090295
- 'Filamentous Fusion Phage: Novel Expression Vectors That Display Cloned Antigens on the Virion Surface', Science (1985), DOI: 10.1126/science.4001944
- 'An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule', PNAS (1967), DOI: 10.1073/pnas.58.1.217





